Allosteric Knot Catalysis - Hic Sunt Dracones!
‘Allosteric Initiation and Regulation of Catalysis with a Molecular Knot’ Vanesa Marcos, Alexander J. Stephens, Javier Jaramillo-Garcia, Alina L. Nussbaumer, Steffen L. Woltering, Alberto Valero, Jean-François Lemonnier, Iñigo J. Vitorica-Yrezabal and David A. Leigh, Science, 352, 1555-1559 (2016). Full Article.
Here be Dragons! The chemistry of molecular knots is an almost completely unexplored area of the molecular world. Now that strategies and tactics are beginning to be developed through which chemists can build such structures, for the first time their properties can be discovered, sometimes with surprising results!

The significance of knotting is increasingly becoming apparent in fields as varied as colloids,1 liquid crystals,2 optical beams,3 soap films,4 superfluids5 and the origins of the early universe6. In molecular terms knots are found in circular DNA7 and approximately 1% of proteins,8 and they form spontaneously in polymer chains of sufficient length and flexibility.9 However, although knots are fundamental elements of structure, the potential benefits that could arise from tying molecules in knots remain mostly unclear.10-13 Now scientists at the University of Manchester have found that as little as 1 mol% of a halophilic synthetic molecular pentafoil knot can promote Lewis acid catalysed reactions by the in situ generation of a carbocation through carbon-halogen bond cleavage induced by the knot (Figure 4).14 The catalysis can be allosterically regulated as the active site of the knot is only formed when it binds five metal(II) ions at bipyridine (bipy) groups that line the outside of—and organise—the halide binding pocket.
Knot Synthesis
A molecular pentafoil knot has previously been formed from a circular metal helicate scaffold through imine chemistry (click here to see this work).15 However, the lability of the imine groups does not allow the metal ions to be removed whilst maintaining the knot topology. So, in order to prepare a wholly organic molecular pentafoil knot, the Leigh group used ring-closing olefin metathesis to assemble a more robust molecular backbone (Figure 1).
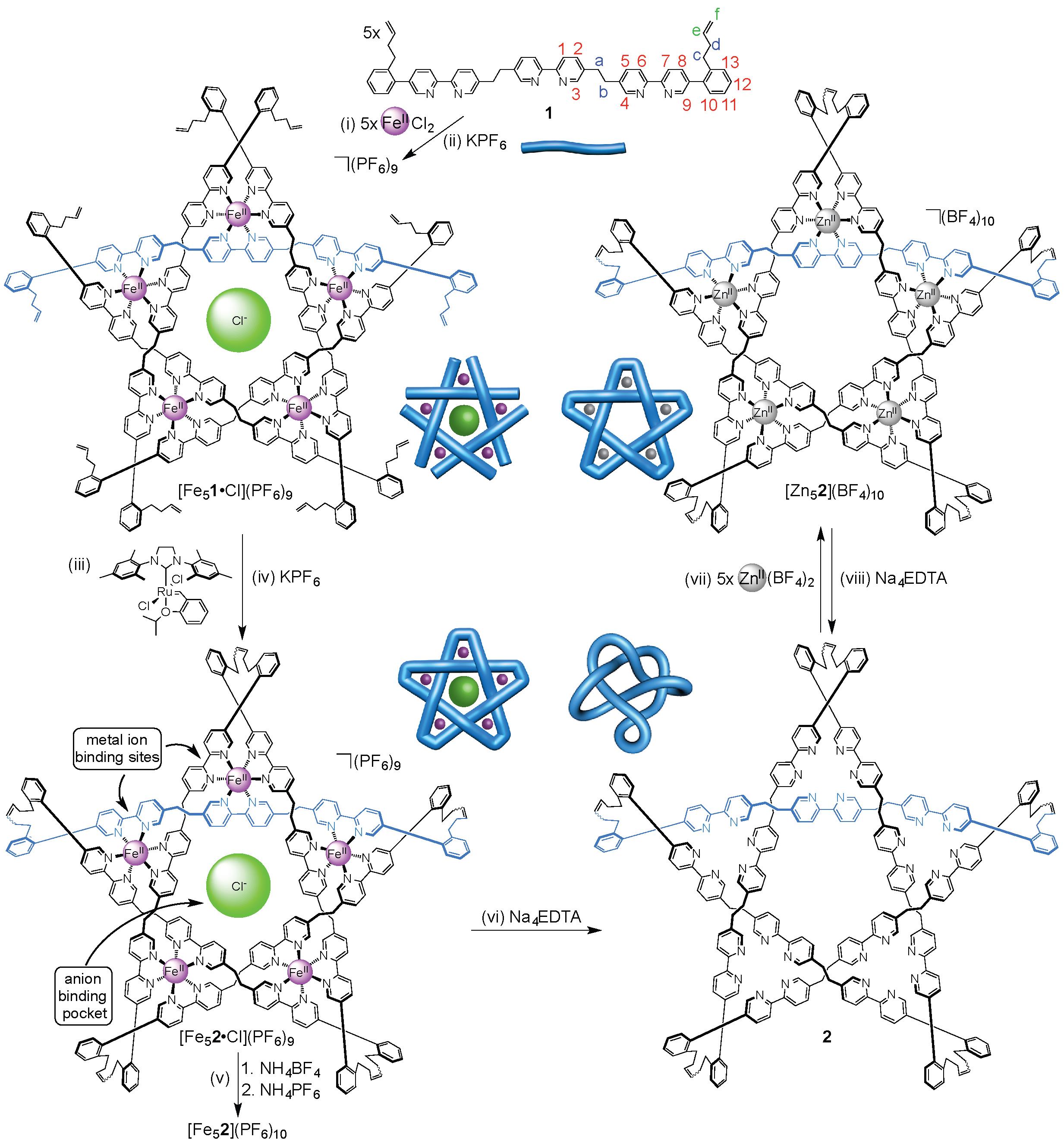
Figure 1. Assembly of wholly organic pentafoil knot 2 and its reversible metallation with zinc(II) ions to form [Zn52](BF4)10.
Unlike the imine knot, the metal ions (and accompanying anions) could be removed from the new pentafoil knot to give a wholly organic pentafoil knot (Figure 1, step vi). The knotted ligand could be remetallated with a different metal ion, Zn(II), under mild conditions (Figure 1, step vii) to give the Zn(II)5-pentafoil knot.
Knot characterization and properties
The X-ray crystal structure of the Fe(II)5-coordinated pentafoil knot (Figure 2) confirms its topology and shows a chloride ion bound tightly in the knot cavity. Indeed, the empty cavity Fe(II)- or Zn(II)-metallated knots are amongst the strongest noncovalent binding synthetic hosts of Cl- and Br- known, readily sequestering traces of chloride from the environment, including solvents and glassware.
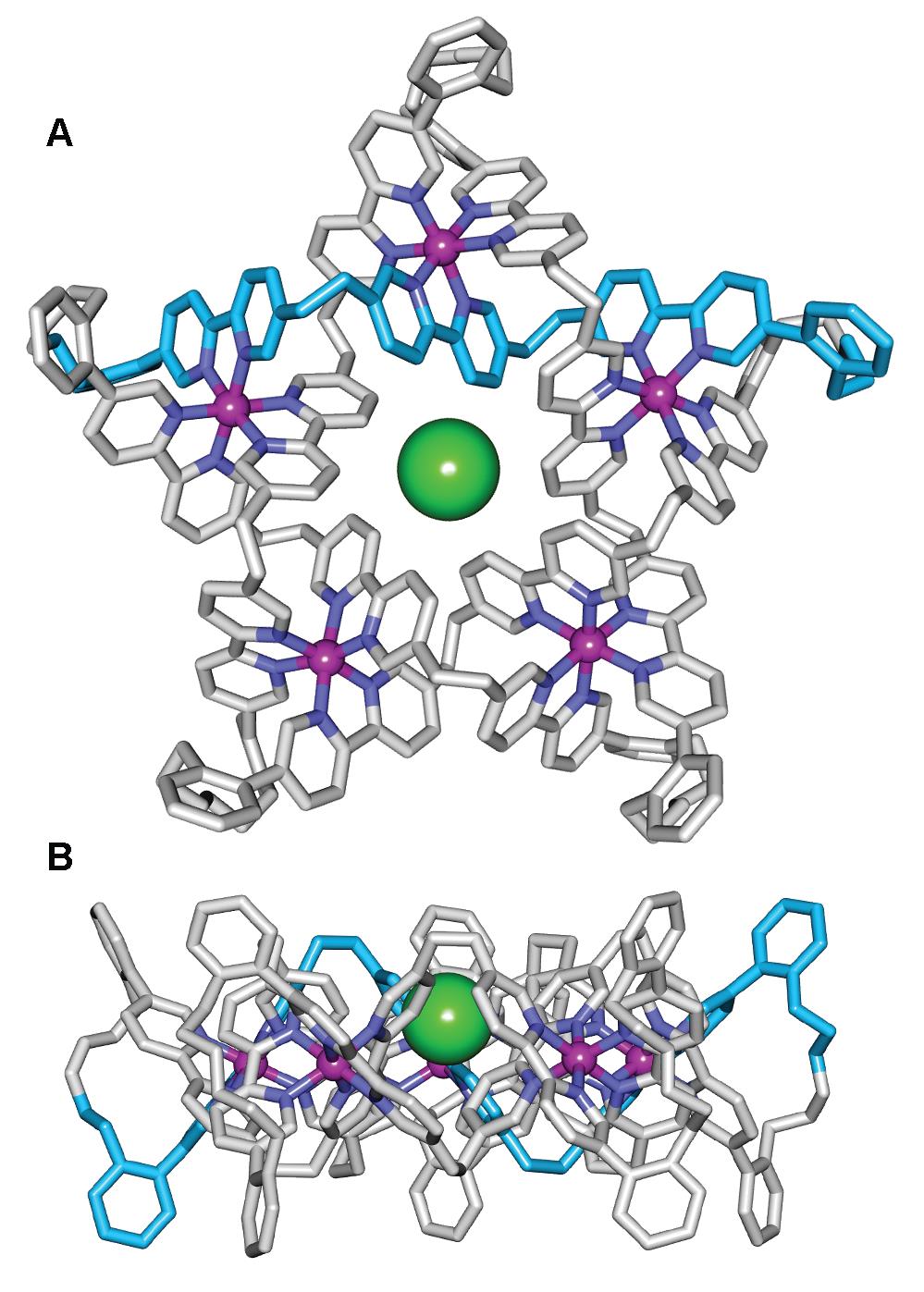
Figure 2. X-Ray crystal structure of pentafoil knot [Fe52•Cl](PF6)6(BPh4)3, shown as the Λ5- enantiomer (Δ5-[Fe52•Cl](PF6)6(BPh4)3 is present in equal amounts in the unit cell). (A) Viewed from above the central cavity of the knot. (B) Viewed in the plane of the Fe(II) cations.
Despite possessing no elements of Euclidean chirality, the metal-free knot is intrinsically chiral by virtue of its topology. The enantiomers of the demetallated pentafoil knot could be separated by HPLC and the pentafoil knots of opposite handedness shown to have circular dichroism (CD) spectra of equal and opposite shape and sign (Figure 3).
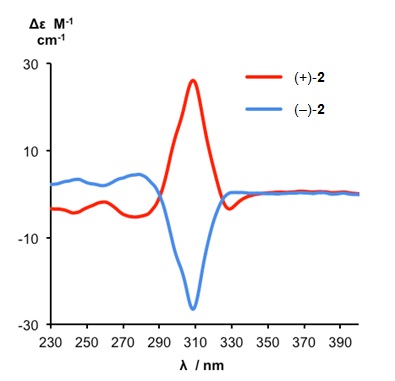
Figure 3. CD spectra of metal-free pentafoil knot enantiomers (+)-2 and (-)-2 (1.26 x 10-4 M, CH2Cl2, 298 K). The pentafoil knot enantiomers are labeled as (+)-2 and (-)-2 according to their specific rotations.
Knot catalysis
The powerful halide binding property of the metallated pentafoil knots could be exploited in anion binding catalysis (Figure 4A). Methyl triflate (MeOTf) reacts with Br- ions bound in the knot cavity to generate MeBr and the empty cavity knot. In this way the knot can catalyse the hydrolysis of Ph2CHBr to Ph2CHOH (Figure 4A).
The knot can also be used to generate other catalytic species (Figure 4B). The powerful anion binding of the empty cavity pentafoil knot can be used to generate a trityl carbocation which, in turn, can act as a catalyst for Michael addition and Diels-Alder reactions (Figure 4B). As the anion binding pocket is only generated from the coordination of the knotted ligand to five metal ions, this is a dramatic example of allostery: On its own the very flexible wholly organic pentafoil knot does not generate carbocations from trityl bromide and cannot promote Diels-Alder or Michael addition reactions. However, in the presence of Zn(II), the knotted ligand binds to five of these metal ions to form a well-ordered, precise, 3D structure with a powerful halide binding site in the central cavity of the knot (Figure 4B, bottom). This promotes halogen-carbon bond fission, in much the same way as occurs in some halogenase enzymes, and the resulting carbocations catalyze the Diels-Alder and Michael addition reactions.
The knot architecture is crucial to the catalysis; unknotted species are catalytically inactive. The results suggest that knotting molecules may be a useful strategy for reducing the degrees of freedom of flexible chains, enabling them to adopt what are otherwise thermodynamically inaccessible conformations. By analogy it may be that a biological role of knotting is to prevent proteins from adopting low energy, but inactive, folded states.
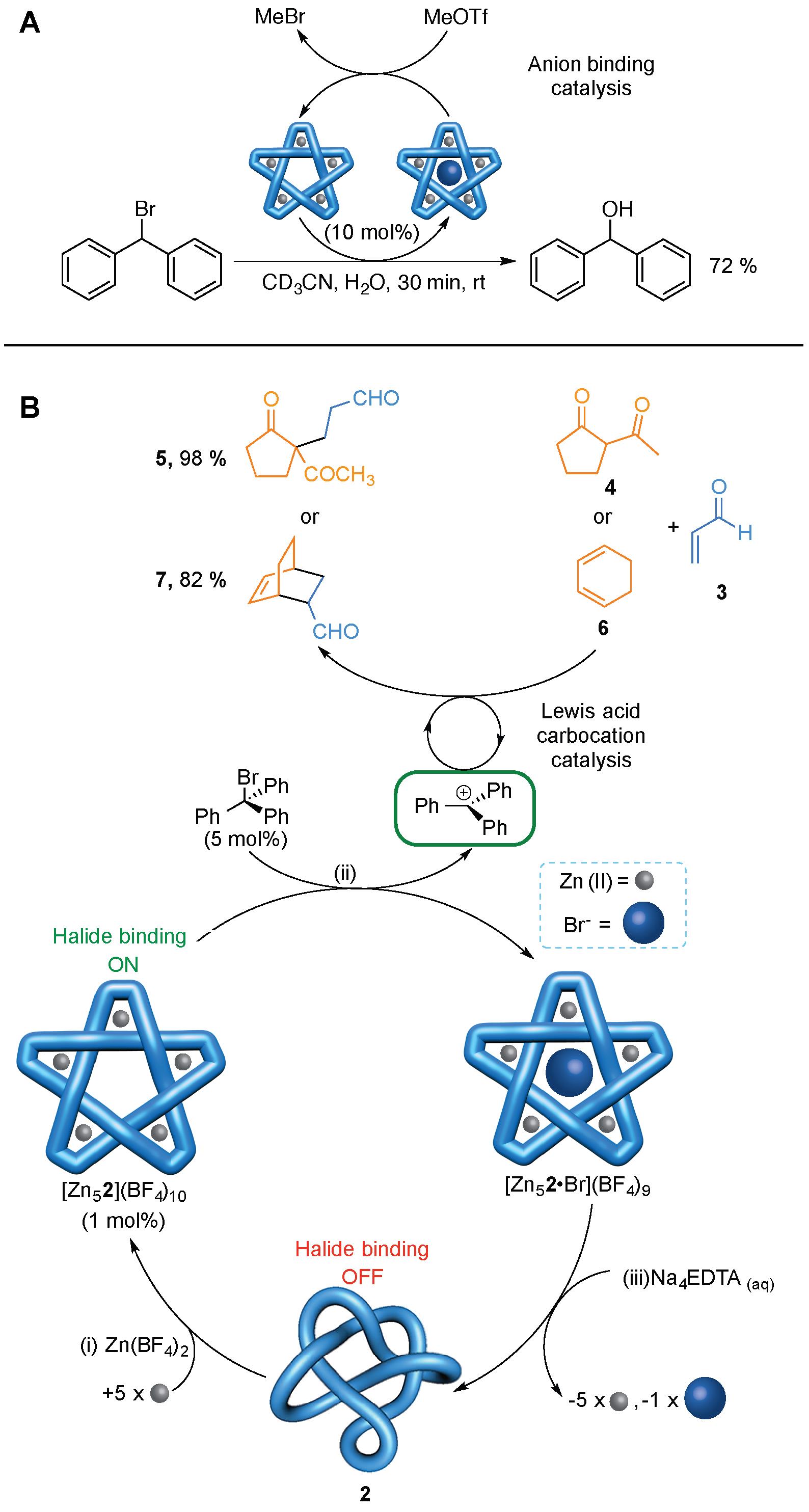
Figure 4. Catalysis using a molecular knot. (A) Catalytic hydrolysis of Ph2CHBr. (B) Allosterically initiated catalysis of Michael addition and Diels-Alder reactions by in situ generation of a trityl cation.
References
[1] J. D. Farrell, C. Lines, J. J. Shepherd, D. Chakrabarti, M. A. Miller and D. J. Wales ‘Structure and stability of charged clusters’, Soft Matter 9, 5407-5416 (2013).
[2] U. Tkalec, M. Ravnik, S. Copar, S. Zumer and I. Musevic ‘Reconfigurable knots and links in chiral nematic colloids’, Science 333, 62-65 (2011).
[3] M. R. Dennis, R. P. King, B. Jack, K. O’Holleran and M. J. Padgett ‘Isolated optical vortex knots’, Nature Phys. 6, 118-121 (2010).
[4] R. E. Goldstein, H. K. Moffatt, A. I. Pesci and R. L. Ricca ‘Soap-film Möbius strip changes topology with a twist singularity’, Proc. Natl. Acad. Sci. U.S.A. 107, 21979-21984 (2010).
[5] D. S. Hall, M. W. Ray, K. Tiurev, E. Ruokokoski, A. H. Gheorghe and M. Möttönen ‘Tying quantum knots’, Nature Phys.(2016) doi:10.1038/nphys3624.
[6] A. Berera, R. V. Buniy, T. W. Kephart, H. Päs and J. G. Rosa ‘Knotty inflation and the dimensionality of spacetime.’, arXiv:1508.01458 [hep-ph]
[7] S. A. Wasserman and N. R. Cozzarelli ‘Biochemical topology: applications to DNA recombination and replication’, Science 232, 951-960 (1986).
[8] J. I. Sułkowska, E. J. Rawdon, K. C. Millett, J. N. Onuchic and A. Stasiak ‘Conservation of complex knotting and slipknotting patterns in proteins’, Proc. Natl. Acad. Sci. U.S.A. 109, E1715-E1723 (2012).
[9] M. D. Frank-Kamenetskii, A. V. Lukashin and A. V. Vologodskii ‘Statistical mechanics and topology of polymer chains’, Nature 258, 398-402 (1975).
[10] A. M. Saitta, P. D. Soper, E. Wasserman and M. L. Klein ‘Influence of a knot on the strength of a polymer strand’, Nature 399, 46-48 (1999).
[11] Y. Arai, R. Yasuda, K.-I. Akashi, Y. Harada, H. Miyata and K. Kinosita Jr., H. Itoh ‘Tying a molecular knot with optical tweezers’, Nature 399, 446-448 (1999).
[12] W. R. Taylor ‘A deeply knotted protein structure and how it might fold’, Nature 406, 916-919 (2000).
[13] J. R. Wagner, J. S. Brunzelle, K. T. Forest and R. D. Vierstra ‘A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome’, Nature 438, 325-331 (2005).
[14] V. Marcos, A. J. Stephens, J. Jaramillo-Garcia, A. L. Nussbaumer, S. L. Woltering, A. Valero, J.-F. Lemonnier, I. J. Vitorica-Yrezabal and D. A. Leigh ‘Allosteric initiation and regulation of catalysis with a molecular knot’, Science 352, 1555-1559 (2016).
[15] J.-F. Ayme, J. E. Beves, D. A. Leigh, R. T. McBurney, K. Rissanen and D. Schultz ‘A synthetic molecular pentafoil knot’, Nature Chem. 4, 15-20 (2015).
