Making molecules that make molecules
‘Sequence-Specific Peptide Synthesis by an Artificial Small-Molecule Machine’ B. Lewandowski, G. De Bo, J. W. Ward, M. Papmeyer, S. Kuschel, M. J. Aldegunde, P. M. E. Gramlich, D. Heckmann, S. M. Goldup, D. M. D’Souza, A. E. Fernandes and D. A. Leigh, Science, 339, 189-193 (2013). Full Article. News & Views. Video.
Tools for a molecular factory: A molecule that makes molecules [Image credit: Guillaume De Bo]
Nature builds proteins in complex molecular factories where information from the genetic code is used to programme the linking of molecular building blocks in the correct order.1 The most extraordinary of these factories is the ribosome,2 a massive molecular machine found in all living cells that assembles amino acids from transfer RNA (tRNA) building blocks into a peptide chain with an order defined by the sequence of the messenger RNA (mRNA) strand that the molecular machine moves along.
Now Professor David Leigh’s group at the University of Manchester (www.catenane.net) have built an artificial molecular machine that builds chemical structures in a similar way.3 Their molecular machine features a functionalized nanometer-sized ring that moves along a molecular track, picking up building blocks located on the path and connecting them together in a specific order to synthesize the desired new molecule.
The mechanism of operation of the molecular machine is shown in Figure 1 (and is also shown in a video). First the ring is threaded onto a molecular strand using copper ions to direct the assembly process. Then a “reactive arm” is attached and the machine starts to operate. The ring moves up and down the strand until its path is blocked by a bulky group. The reactive arm then detaches the obstruction from the track and transfers it to another site on the machine, regenerating the active site on the arm. The ring is then free to move further along the strand until its path is obstructed by the next building block. This, in turn, is removed and passed to the elongation site on the ring, thus building up a new molecular structure. Once all the building blocks are removed from the track, the ring de-threads and the synthesis is complete.
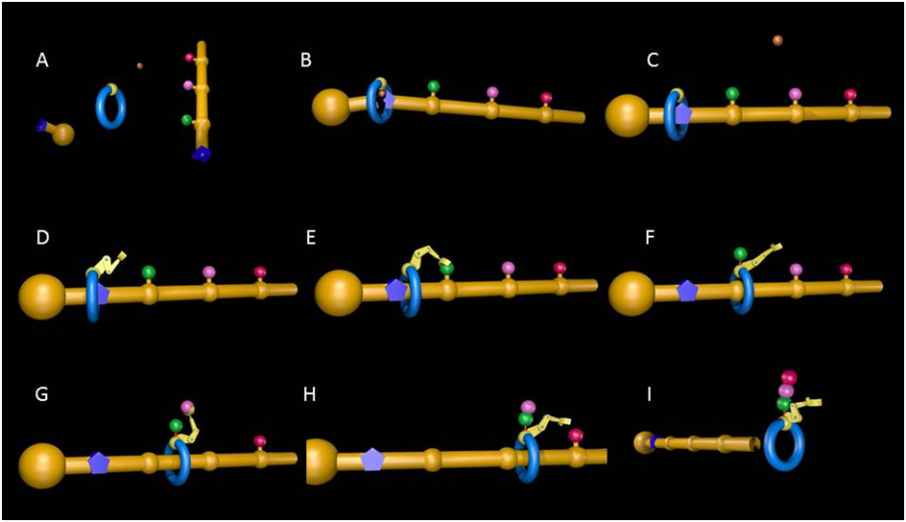
Figure 1. Mechanism of operation of the artificial molecular machine [image credit: Miriam Wilson]. A: Molecular ring, strand with building blocks (green, pink and red spheres) attached, stopper group and a copper ion. B: The copper ion directs the threading of the ring onto the strand and causes a chemical reaction that attaches the stopper, locking the components together (C). D: The reactive ‘arm’ is attached and the machine is now ready for operation. E: Molecular synthesis begins: The arm picks up the first building block (green) from the strand and, F, attaches it to a site on the moving ring. The ring is now free to move to the second building block (pink) which is, in turn, detached from the strand (G) and connected to the green unit (H). I: The process continues until all the building blocks have been removed from the strand by the molecular machine, at which point the synthesis is finished and the ring de-threads with the newly-formed peptide molecule attached (I).
Today the chemical products of the modern world—plastics, paints, pharmaceuticals, catalysts etc—are made by mixing together successive cocktails of reactive chemicals, in processes that are often laborious, inefficient and require many expensive steps. By contrast, in nature molecules are made by other molecules with exquisite efficiency. Biology has not evolved to do this over 2.5 billion years for no good reason and when scientists learn how to use molecular machines to perform synthesis—positioning substrates and ‘reactive arms’ and controlling the dynamics of responsive centres—it will have the potential to revolutionalize the whole approach to functional molecule and materials design.
References
[1] J. M. Berg, J. L. Tymoczko, L. Stryer, Biochemistry (W. H. Freeman, New York, 6th edition, 2006).
[2] A. Yonath, Angew. Chem. Int. Ed. 49, 4340 (2010).
[3] B. Lewandowski, G. De Bo, J. W. Ward, M. Papmeyer, S. Kuschel, M. J. Aldegunde, P. M. E. Gramlich, D. Heckmann, S. M. Goldup, D. M. D’Souza, A. E. Fernandes and D. A. Leigh, Science 339, 189-193 (2013).

