Vernier Entanglements
‘Vernier Template Synthesis of Molecular Knots’ Zoe Ashbridge, Elisabeth Kreidt, Lucian Pirvu, Fredrik Schaufelberger, Joakim Halldin Stenlid, Frank Abild-Pedersen & David A. Leigh, Science, 375, 1035-1041 (2022). Full Article.
The entanglement of molecular strands has profound structural consequences: it blocks conformations, alters dynamics and enhances structural stability.1 Consequently, systematic strand entanglements affect the properties and functions of both synthetic2 and biological3 macromolecules, from small knots to large extended entangled arrays such as bacteriophage capsids4 and the kinetoplast5. Relatively small molecular knots are synthetically accessible6 through, for example, cyclic helicate7 scaffolds and template chemistry,8 but such synthetic approaches have a practical size limit. Until now there have been no strategies for synthesizing the more sizeable arrays of systematic molecular entanglements necessary for larger knotted structures and assemblies. Now the Leigh group report an effective means of rapidly and efficiently assembling such structures from small building blocks using Vernier templating.
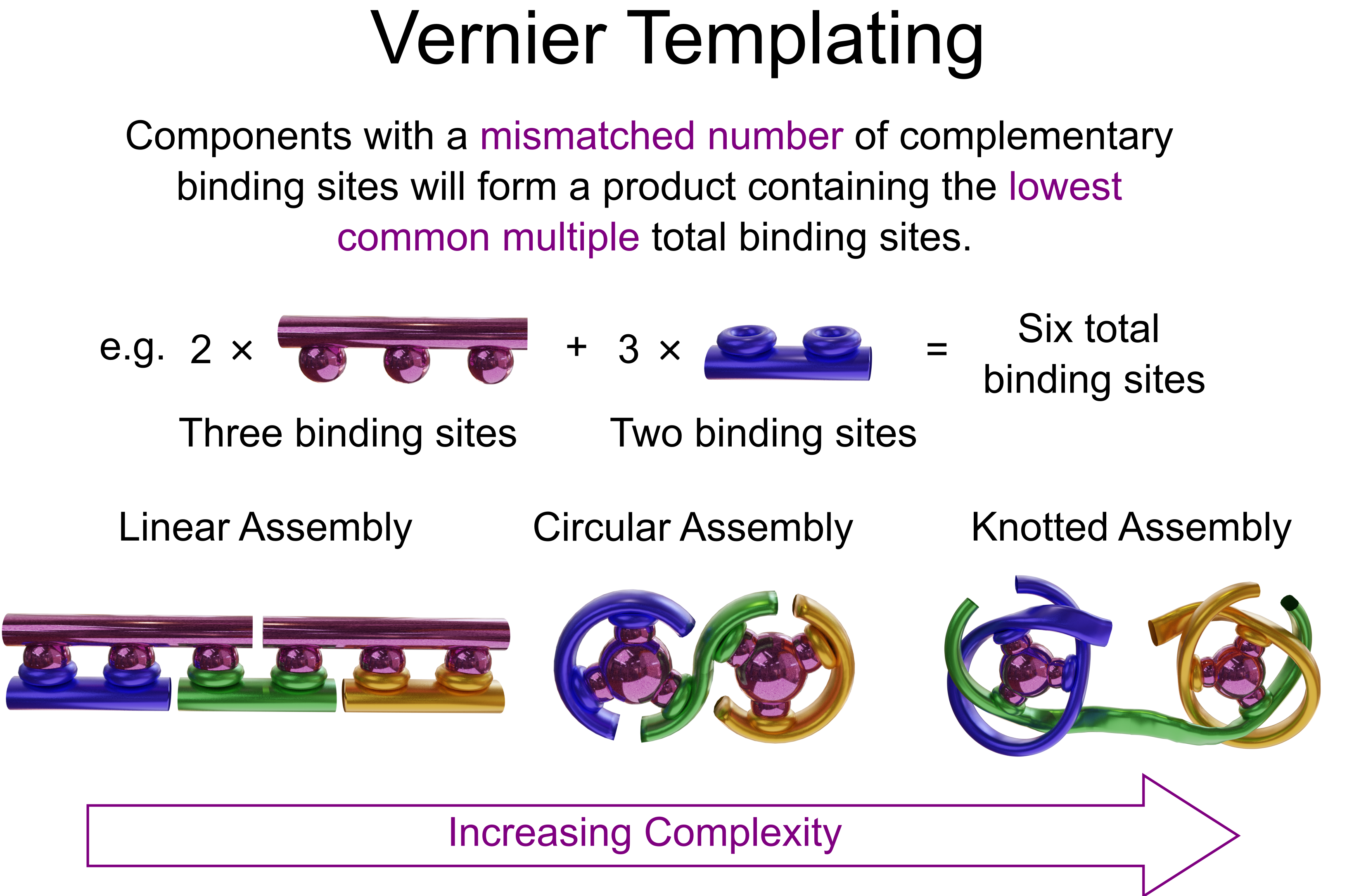
Vernier templating relies on a mismatch between the number of binding sites on two complementary components (Fig. 1).9 The result is a Vernier complex with the lowest common multiple of binding sites of the two components. This approach has previously been used to assemble very large, but topologically trivial, macrocycles (Fig. 1).10
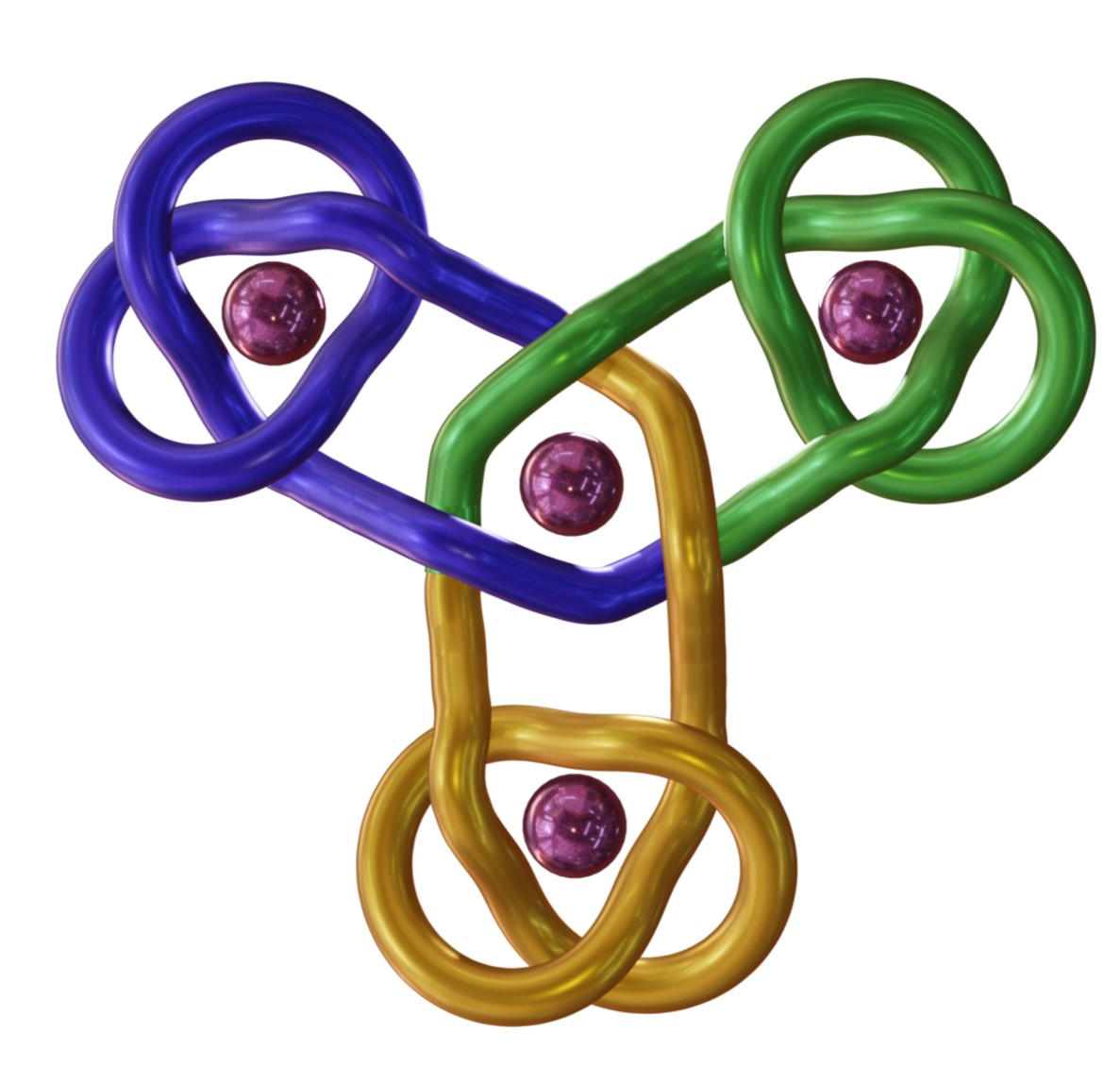
The Leigh group has now adapted the Vernier concept to the assembly of molecular knots by complexing ligand strands with two or four tridentate pyridinedicarboxamide (pdc) groups with nine-coordinate lanthanide ions: A 3:4 (tetratopic strand:metal) Vernier complex gives a 12-crossing triskelion (trefoil-of-trefoils) knot with complete topological stereocontrol (Fig. 2).11
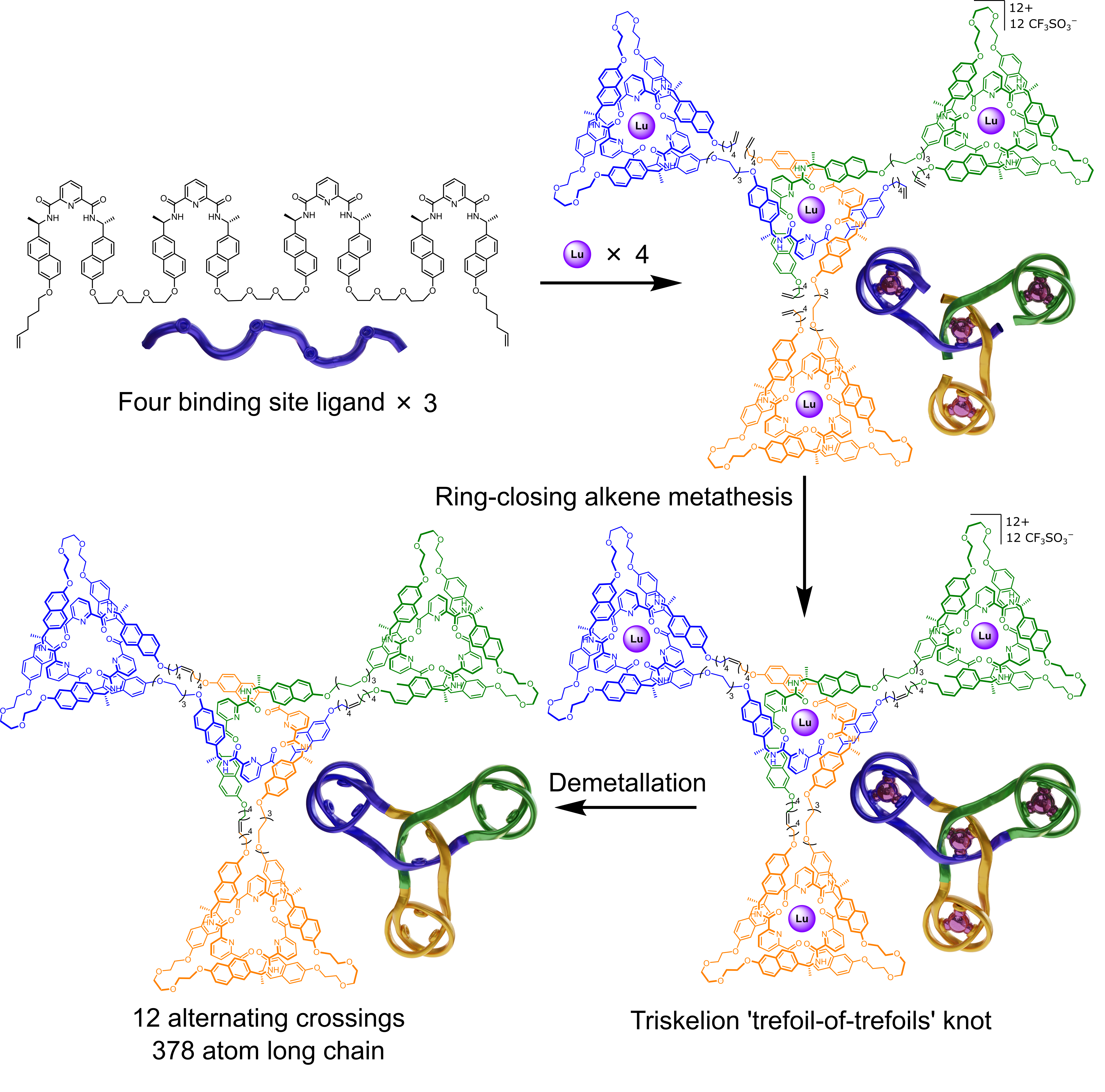
Furthermore, because one chiral handedness of pdc unit cannot coordinate to a lanthanide ion that is coordinated to a pdc of opposite handedness, a new selection element could be introduced into Vernier templating. While a tetratopic ligand strand with all (R)-chiral pdc groups affords a triskelion knot with a topology of 12 alternating crossings, a tetratopic ligand strand with 3 sets of (R)-pdc groups and one (S)-pdc group forms a Vernier complex, and subsequent triskelion knot, with an inverted central core (Fig. 3).11 The inverted-core triskelion knot has 6 alternating crossings and 6 non-alternating crossings.

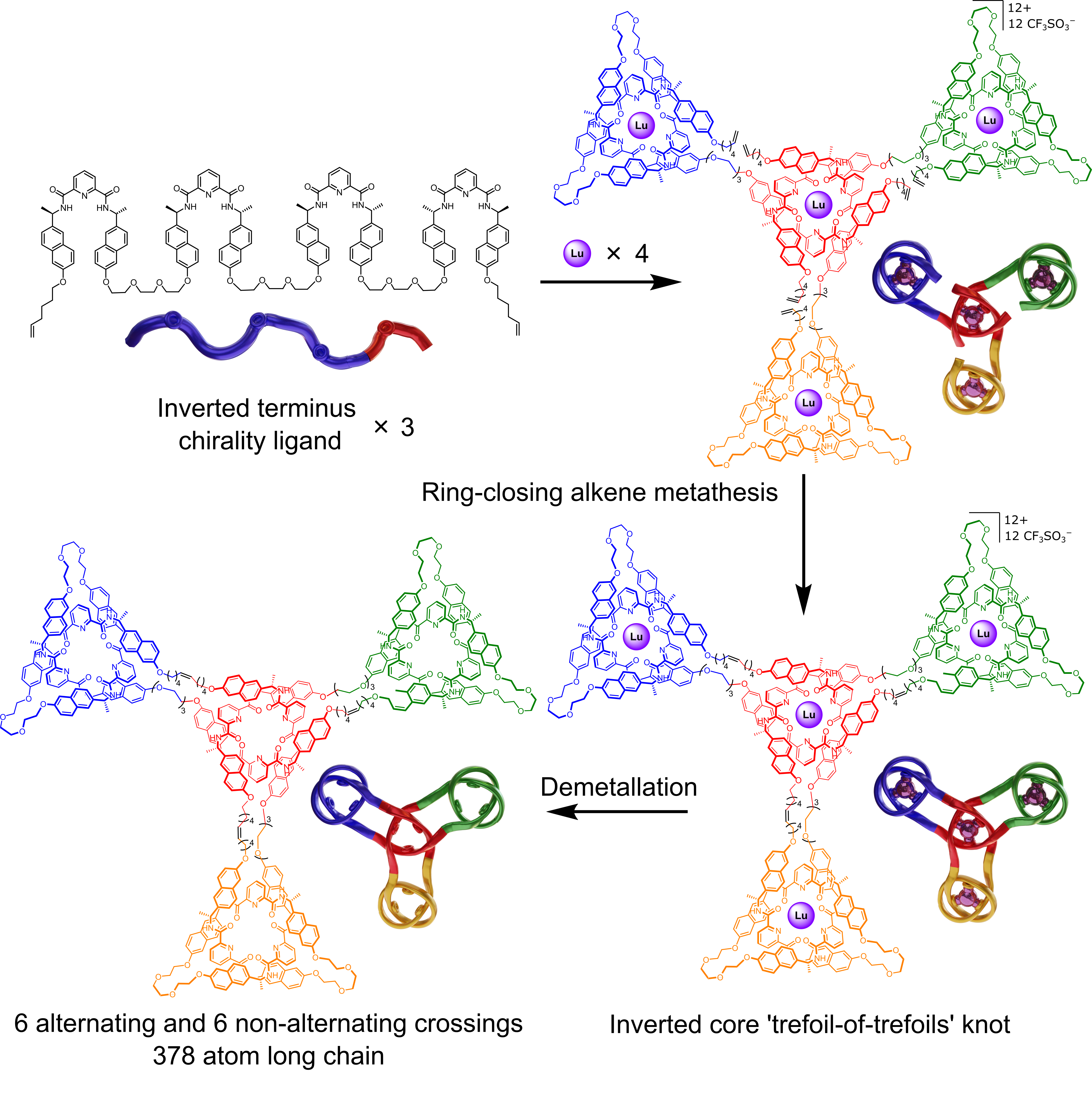
The triskelion and inverted-core triskelion knots are the largest, and most topologically complex, discrete arrays of molecular entanglements prepared to date. The strands are 40 nm, 378-atom-long strands, the same length as a 126-residue peptide, and are essentially two-dimensional. Knotting is conceptually related to weaving:12 materials consisting of 2D/3D arrays of multiple strands that have periodic mutual entanglement. The large entangled arrays available through Vernier templating are more structurally related to materials produced by knitting. That is they consist of a single 1D strand (not multiple, mutually entangled, strands), that is systematically self-entangled to form a persistent 2D layer.13
The unavailability of all but the simplest types of molecular knots hinders the investigation of the effects of systematically entangled structures. The ability to synthesize large, hierarchically knotted, molecular architectures by Vernier templating—with precise control of entanglements, crossing stereochemistry and overall symmetry—opens up new opportunities and research directions for topological molecules and materials.

References
1. S. D. P. Fielden, D. A. Leigh, S. L. Woltering, Molecular knots. Angew. Chem. Int. Ed. 56, 11166–11194 (2017).
2. J. F. Stoddart, Dawning of the age of molecular nanotopology. Nano Lett. 20, 5597–5600 (2020).
3. N. C. H. Lim, S. E. Jackson, Molecular knots in biology and chemistry. J. Phys.: Condens. Matter 27, 354101 (2015).
4. W. R. Wikoff, L. Liljas, R. L. Duda, H. Tsuruta, R. W. Hendrix, J. E. Johnson, Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289, 2129–2133 (2000).
5. J. Chen, C. A. Rauch, J. H. White, P. T. Englund, N. R. Cozzarelli. The topology of the kinetoplast DNA network. Cell 80, 61–69 (1995).
6. (a) C. O. Dietrich-Buchecker, J.-P. Sauvage, A synthetic molecular trefoil knot. Angew. Chem. Int. Ed. 28, 189–192 (1989). (b) J. Brüggemann, S. Bitter, S. Müller, W. M. Müller, U. Müller, N. M. Maier, W. Lindner, F. Vögtle, Spontaneous knotting—From oligoamide threads to trefoil knots. Angew. Chem. Int. Ed. 46, 254–259 (2007). (c) N. Ponnuswamy, F. B. L. Cougnon, J. M. Clough, G. D. Pantoş, J. K. M. Sanders, Discovery of an organic trefoil knot. Science 338, 783–785 (2012). (d) F. B. L. Cougnon, K. Caprice, M. Pupier, A. Bauza, A. Frontera, A strategy to synthesize molecular knots and links using the hydrophobic effect. J. Am. Chem. Soc. 140, 12442–12450 (2018). (e) Y. Segawa, M. Kuwayama, Y. Hijikata, M. Fushimi, T. Nishihara, J. Pirillo, J. Shirasaki, N. Kubota, K. Itami, Topological molecular nanocarbons: All-benzene catenane and trefoil knot. Science 365, 272–276 (2019). (f) H.-N. Zhang, W.-X. Gao, Y.-J. Lin, G.-X. Jin, Reversibly structural transformation between a molecular Solomon link and an unusual unsymmetrical trefoil knot. J. Am. Chem. Soc. 141, 16057–16063 (2019). (g) L.-L. Dang, H.-J. Feng, Y.-J. Lin, G.-X. Jin, Self-assembly of molecular figure-eight knots induced by quadruple stacking interactions. J. Am. Chem. Soc. 142, 18946–18954 (2020).
7. (a) J.-F. Ayme, J. E. Beves, D. A. Leigh, R. T. McBurney, K. Rissanen, D. Schultz, A synthetic molecular pentafoil knot. Nat. Chem. 4, 15–20 (2012). (b) R. A. Bilbeisi, T. Prakasam, M. Lusi, R. El-Khoury, C. Platas-Iglesias, L. J. Charbonnière, J.-C. Olsen, M. Elhabiri, A. Trabolsi, [C–H···anion] interactions mediate the templation and anion binding properties of topologically non-trivial metal-organic structures in aqueous solutions. Chem. Sci. 7, 2524−2532 (2016). (c) J. J. Danon, A. Krüger, D. A. Leigh, J.-F. Lemonnier, A. J. Stephens, I. J. Vitorica-Yrezabal, S. L. Woltering, Braiding a molecular knot with eight crossings. Science 355, 159–162 (2017). (d) Y. Inomata, T. Sawada, M. Fujita, Metal-peptide torus knots from flexible short peptides. Chem 6, 294–303 (2020). (e) Y. Inomata, T. Sawada, M. Fujita, Metal−peptide nonafoil knots and decafoil supercoils. J. Am. Chem. Soc. 143, 16734–16739 (2021).
8. (a) H. Adams, E. Ashworth, G. A. Breault, J. Guo, C. A. Hunter, P. C. Mayers, Knot tied around an octahedral metal centre. Nature 411, 763 (2001). (b) G. Gil-Ramírez, S. Hoekman, M. O. Kitching, D. A. Leigh, I. J. Vitorica-Yrezabal, G. Zhang, Tying a molecular overhand knot of single handedness and asymmetric catalysis with the corresponding pseudo-D3-symmetric trefoil knot. J. Am. Chem. Soc. 138, 13159–13162 (2016). (c) D. A. Leigh, J. J. Danon, S. D. P. Fielden, J.-F. Lemonnier, G. F. S. Whitehead, S. L. Woltering, A molecular endless (74) knot. Nat. Chem. 13, 117–122 (2021). (d) J. P. Carpenter, C. T. McTernan, J. L. Greenfield, R. Lavendomme, T. K. Ronson, J. R. Nitschke, Controlling the shape and chirality of an eight-crossing molecular knot. Chem 7, 1407–1409 (2021).
9. (a) J. S. Lindsey, Self-assembly in synthetic routes to molecular devices. Biological principles and chemical perspectives: A review. New J. Chem. 15, 153–180 (1991). (b) T. R. Kelly, R. L. Xie, C. Kraebel Weinreb, T. Bregant, A molecular Vernier. Tetrahedron Lett. 39, 3675–3678 (1998). (c) C. A. Hunter, S. Tomas, Accurate length control of supramolecular oligomerization: Vernier assemblies. J. Am. Chem. Soc. 128, 8975–8979 (2006). (d) X. Li, C. Hao, C. Tian, P. Wang, C. Mao, Vernier assembly: controlling DNA polymerization via length mismatching. Chem. Commun. 50, 6361–6363 (2014). (e) T. Wei, J. H. Jung, T. F. Scott, Dynamic covalent assembly of peptoid-based ladder oligomers by Vernier templating. J. Am. Chem. Soc. 137, 16196–16202 (2015).
10. (a) M. C. O’Sullivan, J. K. Sprafke, D. V. Kondratuk, C. Rinfray, T. D. W. Claridge, A. Saywell, M. O. Blunt, J. N. O’Shea, P. H. Beton, M. Malfois, H. L. Anderson, Vernier templating and synthesis of a 12-porphyrin nano-ring. Nature 469, 72–75 (2011). (b) D. V. Kondratuk, L. M. A. Perdigao, M. C. O'Sullivan, S. Svatek, G. Smith, J. N. O'Shea, P. H. Beton, H. L. Anderson, Two Vernier-templated routes to a 24-porphyrin nanoring. Angew. Chem. Int. Ed. 51, 6696–6699 (2012). (c) D. V. Kondratuk, J. K. Sprafke, M. C. O'Sullivan, L. M. A. Perdigao, A. Saywell, M. Malfois, J. N. O'Shea, P. H. Beton, A. L. Thompson, H. L. Anderson, Vernier-templated synthesis, crystal structure, and supramolecular chemistry of a 12-porphyrin nanoring. Chem.-Eur. J. 20, 12826–12834 (2014).
11. Z. Ashbridge, E. Kreidt, L. Pirvu, F. Schaufelberger, J. Halldin Stenlid, F. Abild-Pedersen, D. A. Leigh, Vernier template synthesis of molecular knots. Science, 375, 1035-1041 (2022).
12. Z.-H. Zhang, B. J. Andreassen, D. P. August, D. A. Leigh, L. Zhang, Molecular weaving. Nat. Mater. published online 3 Feb 2022.
13. D. P. August, R. A. W. Dryfe, S. J. Haigh, P. R. C. Kent, D. A. Leigh, J.-F. Lemonnier, Z. Li, C. A. Muryn, L. I. Palmer, Y. Song, G. F. S. Whitehead, R. J. Young, Self-assembly of a layered 2D molecularly woven fabric. Nature 588, 429–435 (2020).
